Research
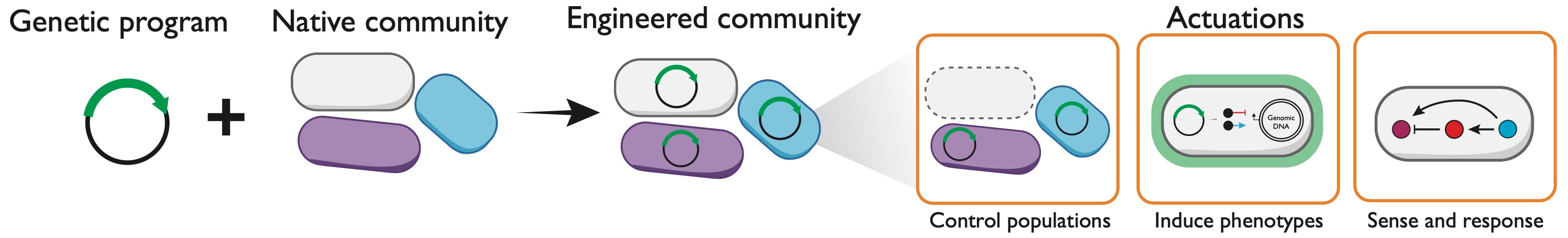
Our group is broadly interested in creating new biotechnologies based upon genetically engineered microbes. Our research focuses on two important challenges toward this goal: (1) advancing our ability to genetically engineer the large numbers of undomesticated, non-model microbes in the laboratory, and (2) creating new technologies to allow microbial consortia to be genetically programmed in their native environment (e.g., gut, soil, marine, wastewater). To achieve this, we use engineered RNA molecules to create memory devices, sensors, and gene regulators to program individual cells and consortia.
Click on the sections below to learn more about our current research areas.
Click on the sections below to learn more about our current research areas.
1. PLUG-AND-PLAY RNA SENSING AND MEMORY SYSTEM
|
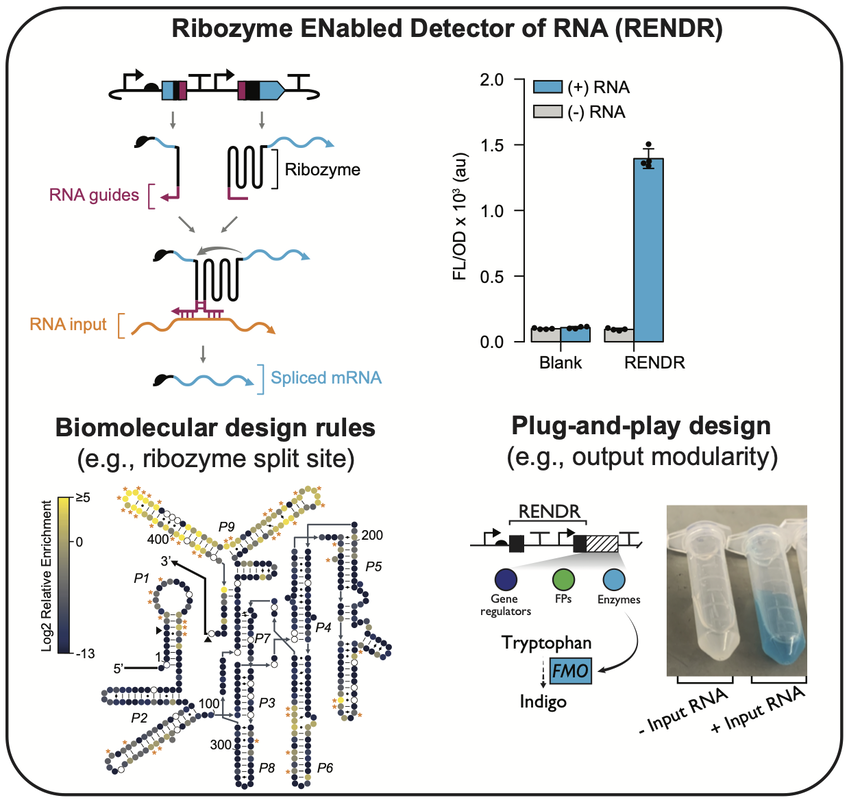
Direct genetic programming of microbial consortia in their native habitats would provide a new capability to profile and engineer microbiomes in situ. This would advance our ability to program microbiomes for therapeutic applications and uncover a deeper understanding of how microbes influence human health. To realize this, we need an understanding of how to design genetic programs that transfer efficiently through microbiomes in situ. Additionally, for certain applications, it will be desirable to control genetic programs to only act in specific community members. Toward this, our lab is creating new technologies based upon engineered splicing ribozymes that serve as memory devices to record gene transfer and sensing devices to achieve spatiotemporal control. The example shown (left) is a plug-and-play RNA transduction platform that uses native host RNA to control the activity of genetic programs. This platform called Ribozyme ENabled Detector of RNA (RENDR) uses a splicing ribozyme split into two non-functional halves that are designed to interact with a host-derived RNA input. When present, the RNA input complements the splicing reaction, allowing RENDR to produce a spliced transcript that can encode any orthogonal protein output. We are exploring applications of RENDR to control genetic programs in response to species and phenotype-specific RNA.
|
2. SYNTHETIC GENE REGULATORS FOR PROGRAMMING GENOMES
|
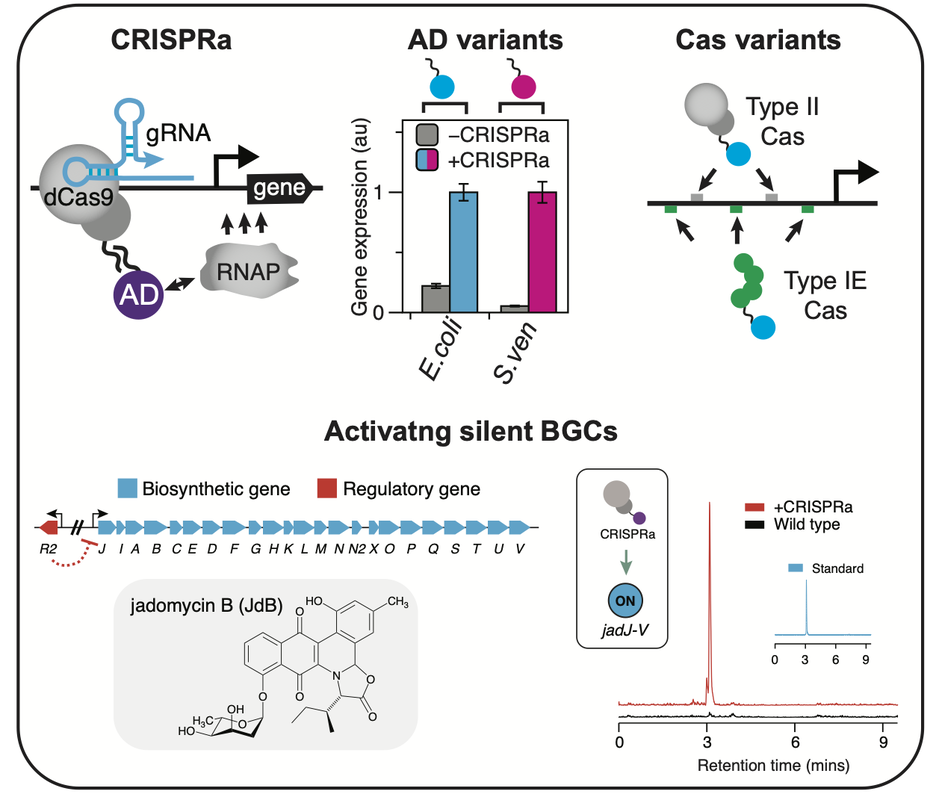
Gene expression is at the heart of how cells control functions and phenotypes. As such, the ability to reprogram how a cell expresses its genome is a potentially transformative capability to uncover gene function or induce valuable phenotypes. We have been creating new tools for reprogramming bacterial gene expression, for example, CRISPR-Cas based activators (CRISPRa) that turn on the transcription of a target gene. Specifically, we have created a highly modular CRISPRa platform where each element of the system (dCas, sgRNA, activation domain [AD]) is independently coded. This allows for facile customization of CRISPRa for different applications. For example, by exchanging the AD, new CRISPRa variants can be created that function in different species. Similarly, we have shown that the dCas component can be interchanged with synthetic or natural Cas proteins, to create new CRISPRa variants that can target different DNA sequences and gene targets.
We are also exploring the applications of CRISPR-Cas regulators, for example, using these systems to activate silent biosynthetic gene clusters (BGC) for drug discovery applications. Our current focus is on applying these technologies in Streptomyces species that are enriched in silent BGC. |
3. SYNTHETIC GENE NETWORKS AND SYNTHETIC CELLS
|
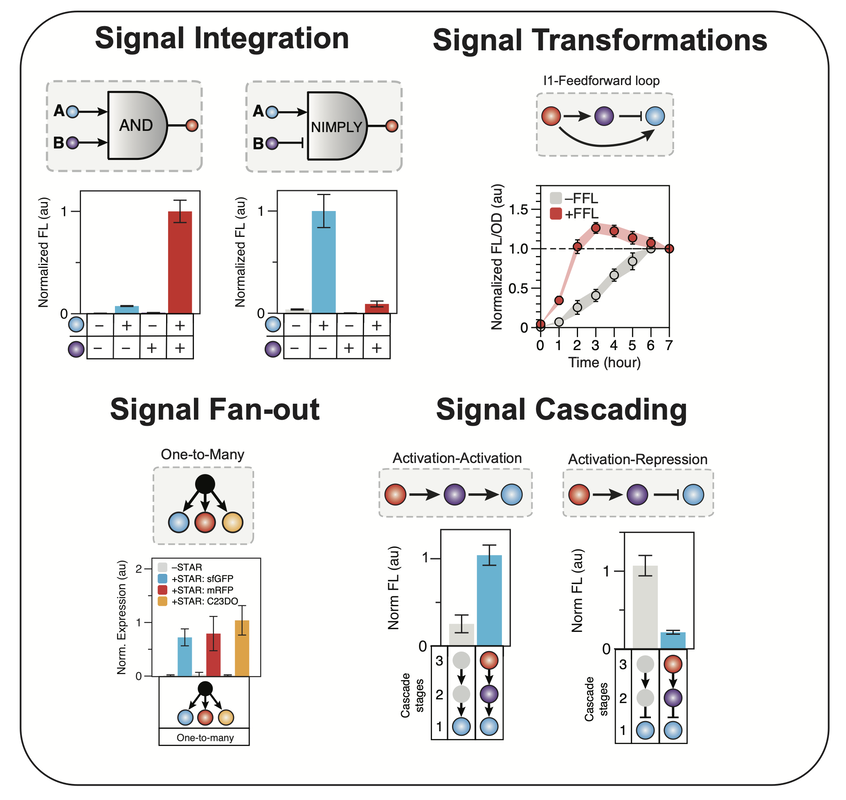
At the heart of how cells make decisions are regulatory networks that allow cells to convert cellular and environmental information into changes in gene expression. Our lab has a long-standing interest in creating and applying synthetic RNA regulators to make synthetic regulatory gene networks. This has largely focused on using synthetic small RNA (sRNA) regulators that include Small Transcription Activating RNA (STAR). We are particularly interested in exploring the broad-host potential of RNA regulators that could allow for the creation of genetic programs that function across diverse microbes and communities of microbes. We are also interested in using synthetic RNA networks for the programming of synthetic cells to replicate life-like functions and enable new applications in biotechnology. |